A Tiny hole – a Huge Breakthrough
Researchers in the Faculty of Biomedical Engineering at the Technion - Israel Institute of Technology have developed a rapid and precise coronavirus test that circumvents PCR amplification errors
Technion Scientists have developed a novel method for rapid and accurate sensing of coronavirus without the need to rely on PCR amplification. The new technique can identify the presence of SARS-CoV-2 in a sample by counting and quantifying the virus’ RNA molecules with single-molecule precision. Sensing is not biased by PCR amplification errors, permitting future development of a more accurate clinical diagnostic technique.

Professor Amit Meller, Dr. Yana Rozevsky, Dr. Xander van Kooten, Dr. Diana Huttner
The research, which was published in ACS Nano, was led by Professor Amit Meller and carried out by postdoctoral researchers Dr. Yana Rozevsky, Dr. Tal Gilboa, Dr. Xander van Kooten, and staff scientist Dr. Diana Huttner – all of whom are researchers in the Faculty of Biomedical Engineering – and Professor Ulrike Stein and Dr. Dennis Kobelt from the Max Delbrück Center for Molecular Medicine and the Charité Hospital in Berlin.
RT-qPCR testing, the most widely used test for COVID-19 today, involves a series of preparatory stages, including collecting the sample from a patient using a swab, “opening” the virus and extracting RNA from it. In the next stage, called reverse transcription (RT), specific ‘target’ RNA sequences are copied to the DNA form. Finally, this DNA is amplified by a polymerase chain reaction (PCR). Millions of copies are made so that enough DNA is present to be detected, finally leading to a diagnosis for COVID-19.
A tiny nanopore device for detecting single biological molecules
RT-qPCR testing requires large quantities of special reagents, expensive laboratory equipment, and highly trained professionals. Moreover, recent studies have shown that test results can change from one day to the next and that the massive amplification process can generate significant errors. For these reasons, worldwide efforts are being devoted to developing faster, more affordable, and more accurate tests. This task is particularly challenging in cases where the “viral load” (the amount of viral RNA) in a sample is low and can evade sensing.
The new method presented by Prof. Meller’s research group relies on original technologies that the lab has developed in the past two decades, using nanofabricated holes (so-called “nanopores”) to sense single biological molecules. The effectiveness of this technology has already been demonstrated in a number of other biomedical uses. Unlike conventional molecular diagnostics, which require large volumes of samples containing millions of copies of the same molecule, nanopore sensing analyzes individual biological molecules from much smaller samples. A strong electrical field is used to unfold and thread individual DNA molecules through the nanoscopic hole containing electrical or optical sensors. Each molecule that passes through the hole gives a characteristic “signature,” which enables identification and immediate counting of the molecules. This approach opens up the possibility of miniaturizing the diagnostic systems while improving the accuracy and reliability of tests and expanding the cases where amplifying PCR is not efficient or harms the reliability of the test.
The recently published article presents two applications of this method: identifying RNA molecules that signal the emergence of metastatic cancer and detecting coronavirus RNA. To allow unambiguous sensing, the researchers developed a process that leaves only the relevant ‘target’ molecules intact, while degrading all others.
In the first application, the researchers demonstrated the method’s potential for early detection of metastatic cancer by quantifying the levels of MACC1 – one of the primary genes known to signal the formation of a metastatic state. Thanks to its high degree of sensitivity, the new technique successfully quantified the gene’s expression in cancerous cells at the early stages of illness (known as stages I and II) – a challenge that PCR-based technologies failed to meet. Needless to say, the earlier these genetic biomarkers are discovered, the better the chances of successful treatment.
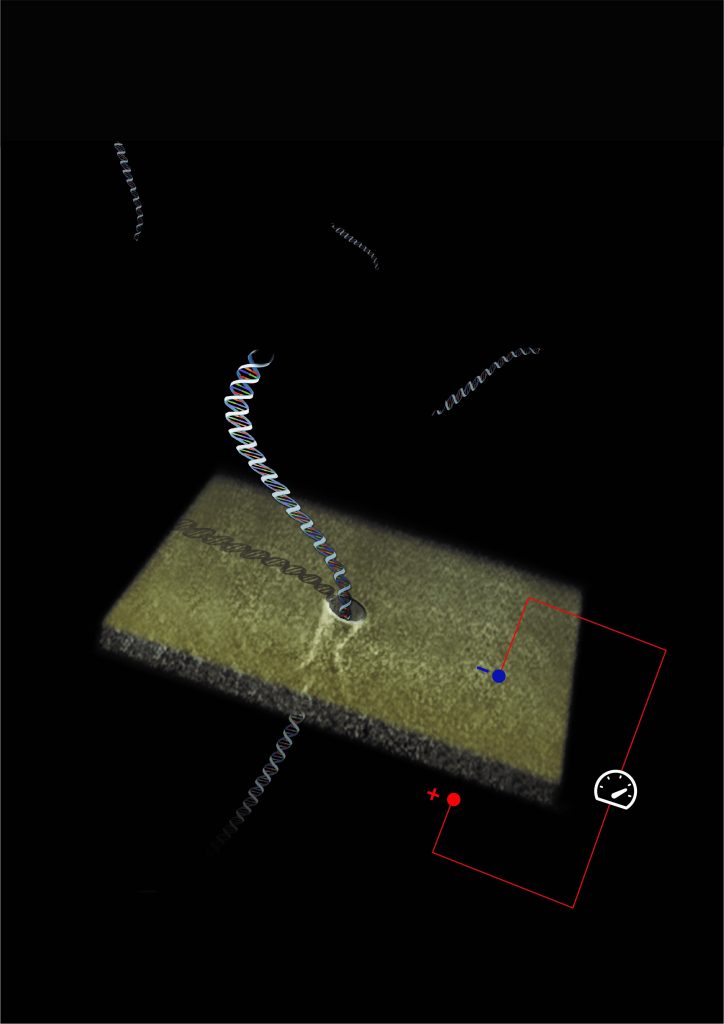
Illustration of DNA molecules passing through a nanopore one after the other
In the second application, the researchers detected the RNA molecules of the SARS-CoV-2 virus using the same approach. The technique presented in the article is not the first to analyze single molecules; however, unlike previous reports, it circumvents processes in the sample treatment that introduce “noise” and errors into the system. Two of these processes are sample purification, in which many target molecules are inadvertently lost, and DNA amplification, which may lead to errors and faulty diagnoses. According to Prof. Meller, “Our system enables quantifiable sensing of the RNA genetic expression levels using a relatively simple nano-sensor device, without needing to cleanse the sample and with no need for massive amplification processes that may harm the test’s sensitivity and reliability. We have shown that our technology preserves the level of genetic expression of the original RNA molecules throughout the entire process. In this way, we obtain a more precise analysis method, which is essential in both contexts we studied – RNA biomarkers of metastatic cancer and the SARS-CoV-2 virus.”
The recent ACS Nano article is an important milestone for Meller’s research group, but it is definitely not the end of the road. With further work, the nanopore sensing system is expected to become a portable device that will make cumbersome lab equipment unnecessary. Technological and clinical research is continuing at the Technion Faculty of Biomedical Engineering, in collaboration with the BioBank at the Rambam Health Care Campus. At the same time, steps are being taken to commercialize the technology in order to make it available for general use as soon as possible.
The research is being supported by the European Union (through an ERC grant, as part of the European Commission’s Horizon 2020 program for research in the EU), the Israel Science Foundation (ISF), and the SignGene program which supports doctoral students.
Click here for the paper in ACS Nano.


